The need for fast simulations is a priority in computational cardiology. Cardiac simulations rely upon heavy computational calculations using finite element methods which currently take a long time on top performing machines. We are implementing graph neural networks to overcome this challenge which have the potential to surrogate finite element solutions. Through applying graph theories, we have developed G-FuNK which can solve electrophysiological differential equations for simulating the atrial electrophysiological dynamics. See the arxiv paper here.

Arrhythmogenic right ventricular cardiomyopathy (ARVC) is a genetic cardiac disease that leads to ventricular tachycardia (VT), a life-threatening heart rhythm disorder. Treating ARVC remains challenging due to the complex underlying arrhythmogenic mechanisms, which involve structural and electrophysiological (EP) remodeling. Here, we developed a novel genotype-specific heart digital twin (Geno-DT) approach to investigate the role of pathophysiological remodeling in sustaining VT reentrant circuits and to predict the VT circuits in ARVC patients of different genotypes. This approach integrates the patient’s disease-induced structural remodeling reconstructed from contrast-enhanced magnetic-resonance imaging and genotype-specific cellular EP properties. In our retrospective study of 16 ARVC patients with two genotypes: plakophilin-2 (PKP2) and gene-elusive (GE), we found that Geno-DT accurately and non-invasively predicted the VT circuit locations for both genotypes (with 100%, 94%, 96% sensitivity, specificity, and accuracy for GE patient group, and 86%, 90%, 89% sensitivity, specificity, and accuracy for PKP2 patient group). This novel Geno-DT approach has the potential to augment therapeutic precision in the clinical setting and lead to more personalized treatment strategies in ARVC. The study was published by eLife in 2023.

Sodium channels play a critical role in the initiation and propagation of the cardiac electrical excitation wavefront. Reductions in the cardiac sodium current impair the action potential upstroke and cardiac conduction, contributing to both acquired and inherited arrhythmia syndromes, such as Brugada syndrome. In recent years, gene therapy has emerged as a promising approach to restore the function of damaged or dysfunctional cells and tissues. In collaboration with Dr. Gerrard Boink’s research team at the University of Amsterdam, we developed a computational framework using human ventricular digital twins to model the delivery of short transcript of SCN10A (S10s) gene therapy for mitigating ventricular arrhythmia . This technology can aid procedural planning for optimal gene therapy injection locations and improve treatment efficacy. Our work has been accepted for publication in the European Heart Journal in January 2025.

Substrate-based ablation is a common therapy for scar-related ventricular tachycardia (VT). Patients are often treated with amiodarone (AMD) prior to the procedure, and there is evidence that this may adversely impact VT recurrence rates by preventing accurate elimination of critical isthmus sites. However, studies disagree on how AMD may alter the distribution of abnormal electrograms (EGMs) used to target these critical sites, and have been unable to fully separate the effects of amiodarone from those of the underlying patient-specific fibrotic substrate. Using personalized computational models, we explored the effects of AMD on substrate mapping. We found that VT inducibility was mildly reduced with addition of AMD, while the area of abnormal signals was significantly increased, limiting the accurate identification of VT isthmus locations. However, when combining EAMs from several pacing locations, regions of isochronal crowding provided a specific marker for critical VT sites that may allow clinicians to mitigate the effects of amiodarone.

Atrial fibrosis is a significant pathophysiological contributor to atrial fibrillation (AF) – the most prevalent cardiac arrhythmia worldwide – and cardiac MRI is the gold standard for assessing fibrotic substrate in the atria. Here, we create personalized atrial digital twins using longitudinal MRI scans taken before and after AF ablation to assess how the ablation lesions impact the substrate's arrhythmogenic potential. By analyzing these models, we can evaluate whether the ablation successfully reduced the substrate’s propensity for arrhythmias and compare substrate characteristics between patients who experienced recurrence and those who did not. This approach seeks to enhance our understanding and improve treatment outcomes for AF patients.
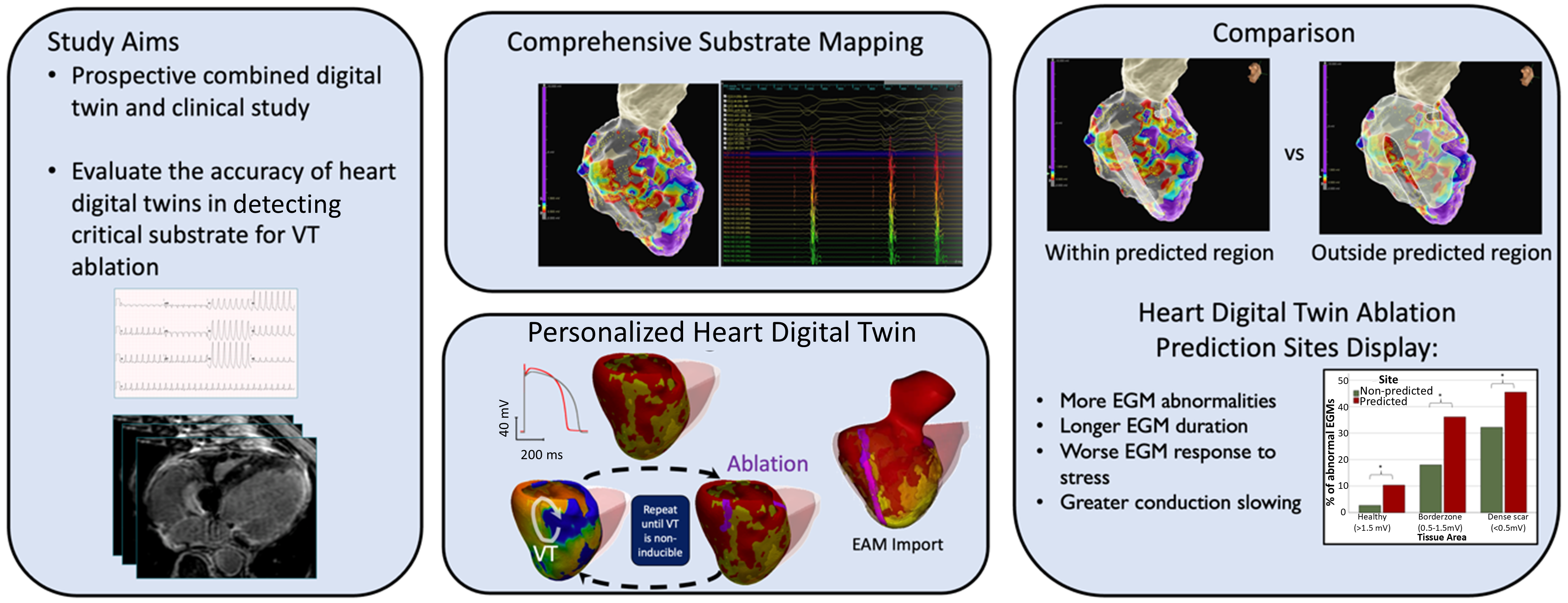
We presented the first combined prospective clinical and digital twin study with the aim of invasively evaluating the accuracy of the personalized heart digital twin predictions. During each VT clinical ablation procedure, we performed comprehensive substrate mapping and evaluated electrogram properties at sites deemed of critical importance by the digital twin in order to establish their relationto the conventional substrate-mapping approach to VT ablation. Heart digital twin–predicted sites display a higher prevalence of abnormal and prolonged electrograms compared with non-predicted sites and accurately identify regions of conduction slowing. This technology may improve the efficacy and act as a guide for catheter ablation of ventricular tachycardia. This work was performed in collaboration with the St George's University of London, UK and has been accepted for publication in the Circulation journal in January 2025 https://doi.org/10.1161/CIRCULATIONAHA.124.070526 .

Cardiacsarcoidosis (CS) is a rare genetic condition affecting the entirebody. Its cardiac manifestation is associated with an increased riskof lethal ventricular arrhythmias (VA) due to fibrotic remodeling.Fibrosis is traditionally detected using Late Gadolinium Enhancement(LGE) MRI which is an expensive and limited resource. Early andaccurate identification of CS patients needing LGE-MRI is essentialfor patient stratification and primary prevention of VA. This studydemonstrates the clinical potential of a deep learning (DL) modelusing 12-lead ECG and electronic health record (EHR) data to predictfibrosis detectable by LGE-MRI. By enabling non-invasive screening,this approach could guide targeted use of LGE-MRI, improve riskstratification, and optimize treatment strategies for CS patients.Read the abstract here (https://doi.org/10.1016/j.cvdhj.2023.08.006)

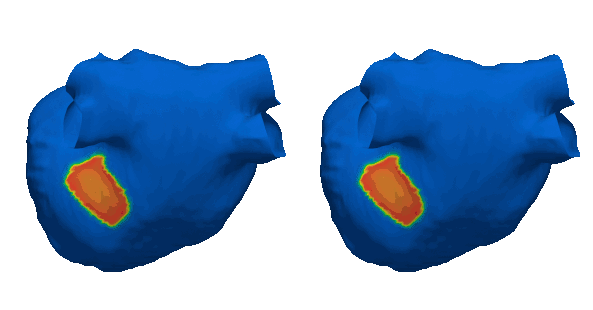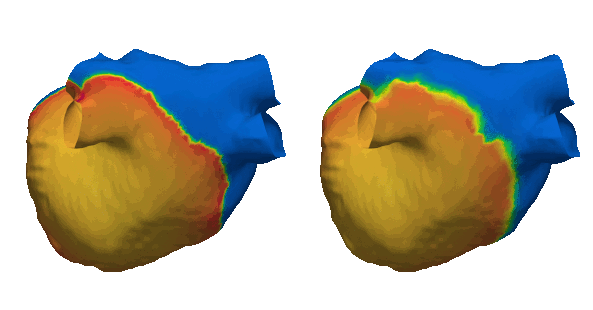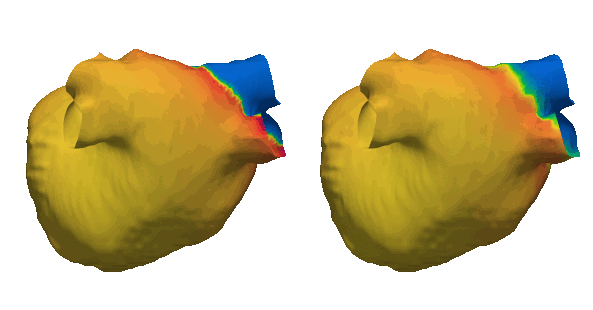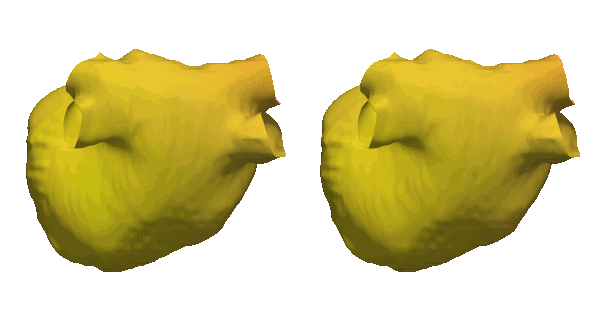
Duchenne muscular dystrophy (DMD) is a rare inherited disease that results in a weakness of muscular tissue, including the heart. As these patients age into adulthood, the weakening of the heart leads to scar formation, cardiac dysfunction, and dangerous ventricular arrhythmias. Currently, there exist no established risk stratification methods to guide clinical decision making in the treatment of ventricular arrhythmias in DMD. By combining digital twin simulations and data-driven analysis, we developed a method that uses standard-of-care MR images alone to non-invasively screen for ventricular arrhythmia risk in patients with DMD.

Although the notion of thrombus formation caused by stasis of blood in the LAA, with subsequent embolism to the brain, was widely accepted to explain the increased risk of stroke in AF, mechanistic insights into why AF patients with certain LAA shapes are more likely to experience embolic events remain elusive. We will develop novel AI [3] in conjunction with mechanistic modeling to investigate the realistic hemodynamics in individualized atrial models and advance our understanding of the mechanisms of how the morphological variations in LAA promote thrombogenesis.
[3] Zan Ahmad, Minglang Yin, et al. A computational pipeline for clustering left atrial appendage morphology via elastic shape analysis. Revised manuscript submitted to Computers in Biology and Medicine.

Numerous studies have applied AI algorithms in clinical prognostication. Due to limited model capabilities, most of these studies have primarily focused on learning from a single data modality. The integration of multimodal data sources and modalities has often been overlooked, which potentially limits AI generalizability in new populations and reduces overall accuracy. The advent of multimodal AI (MAI) provides a promising approach to address this gap by integrating data from multiple sources and modalities.
This theme will entail developing multimodal AI for clinical prognostication. Recently, in a paper submitted to Nature Cardiovascular Research [2], we developed a highly generic MAI framework to predict sudden cardiac death for patients with Hypertrophic Cardiomyopathy. This MAI model is adaptable for various tasks, such as classification, risk stratification, or survival analysis, and is capable of learning from multiple types of data, such as image, clinical covariate, genomic, video, text, or signal. We will actively explore the usage of MAI in clinical prognostication.
[2] Changxin Lai, Minglang Yin, et al. “Multimodal AI to forecast arrhythmic death in hypertrophic cardiomyopathy”. Revised manuscript submitted to Nature Cardiovascular Research
